The Plant Cell 23: 2045-2063 (2011)
A Guideline to Family-wide Comparative State-of-the-art qRT-PCR Analysis Exemplified with a Brassicaceae Cross-species Seed Germination Case Study [W][OA]
* Both authors contributed equally to this work
The University of Nottingham, Division of Statistics, School of Mathematical Sciences, University Park, Nottingham NG7 2RD, United Kingdom (A.T.A.W.)
Received February 8, 2011; revised May 6, 2011; accepted May 27, 2011; published June 10, 2011.
www.plantcell.org/cgi/doi/10.1105/tpc.111.084103
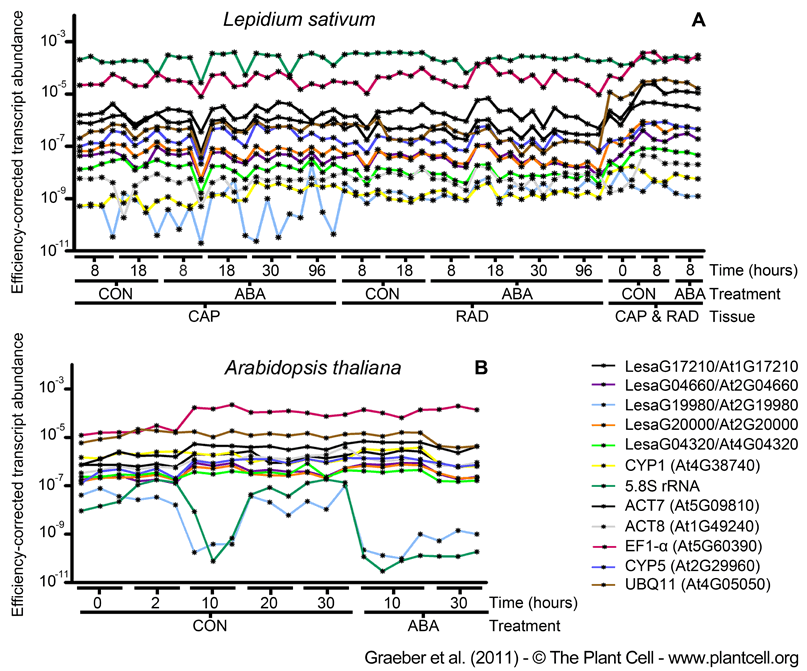
Figure 3. Efficiency-corrected Transcript Abundance of New and Traditionally Used Reference Genes for Brassicaceae Seed Germination as Determined by qRT-PCR.
(A) Lepidium sativum seed tissue RNA samples, CAP (micropylar endosperm) and RAD (radicle plus lower hypocotyl), were dissected from seeds imbibed without (CON) and with ABA added, at the times indicated. In addition, to also consider the dry seed (0h) state for which the two tissues cannot be separated, we also included combined CAP&RAD 0h-samples (and combined 8h-samples for the comparison with the separated 8h-tissues, all combined samples included also testa).
(B) Arabidopsis whole-seed RNA samples from the CON and ABA series. For both species, only seeds prior to the completion of germination, i.e. with unruptured micropylar endosperm, were used for these analyses (as described in Figure 2). PCR efficiency correction was carried out as described in the method section. Single-sample results are presented, but for comparisons and downstream calculations the 3 to 4 biological replicates (as indicated in the graph) can be combined. For new reference genes the L. sativum sequence names and the Arabidopsis AGI codes are indicated. Traditionally used reference genes are indicated with 'trivial name (AGI code)'.
Synopsis: Developmental processes like seed germination are characterised by massive transcriptome changes. This study compares seed transcriptome datasets of different Brassicaceae to identify stable expressed reference genes for cross-species qRT-PCR normalisation. A workflow is presented for improving RNA quality, qRT-PCR performance, and normalisation when analysing expression changes across species.
| Article in PDF format (1.2 MB) Supplemental data file (156 KB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |