The Plant Cell 23: 2045-2063 (2011)
A Guideline to Family-wide Comparative State-of-the-art qRT-PCR Analysis Exemplified with a Brassicaceae Cross-species Seed Germination Case Study [W][OA]
* Both authors contributed equally to this work
The University of Nottingham, Division of Statistics, School of Mathematical Sciences, University Park, Nottingham NG7 2RD, United Kingdom (A.T.A.W.)
Received February 8, 2011; revised May 6, 2011; accepted May 27, 2011; published June 10, 2011.
www.plantcell.org/cgi/doi/10.1105/tpc.111.084103
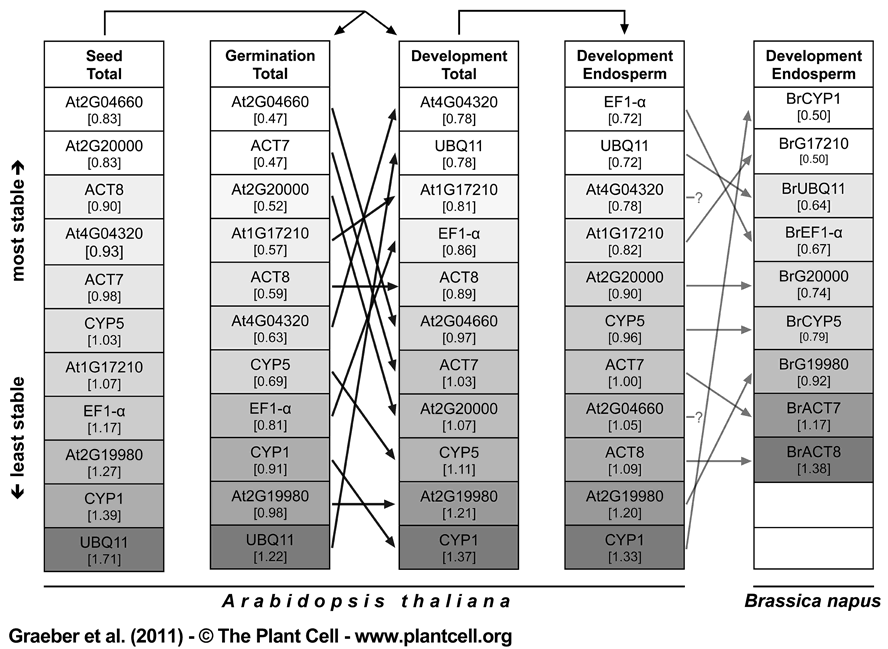
Figure 5. Comparative Gene Expression Stability Analysis of Seed-related Brassicaceae Transcriptome Datasets for Reference Genes Used in the Brassicaceae Cross-species qRT-PCR Seed Germination Study.
For the traditional and new Lepidium sativum and Arabidopsis reference genes (Figure 4) Arabidopsis microarray data of 101 diverse seed-related experiments ('Seed total' dataset) was partitioned into the 'Germination total' dataset (containing only experiments focusing on the seed germination process) and the 'Development total' dataset (containing only experiments focusing on embryogenesis/seed development). The latter was reduced into the 'Development Endosperm' dataset (containing only experiments focusing on endosperm development) indicated by arrows above columns and detailed in methods and Supplemental Dataset 2. Brassica napus microarray data for endosperm development of putative orthologs of the Arabidopsis reference genes was analysed (see methods) and genes named accordingly. Both endosperm datasets consist of highly comparable developmental stages (see methods). Each dataset was analysed by GeNORM to rank the genes by their average expression stability measure M (value in brackets; proportional greyscale colour intensity). Black arrows between columns indicate differences in gene expression stability rankings between the two seed processes, germination and development, for Arabidopsis. Grey arrows between columns indicate differences between the two species Arabidopsis and B. napus for the endosperm development process. The putative B. napus orthologs of Arabidopsis genes At4G04320 and At2G04660 are not present on the Brassica microarray and are therefore left blank.
Synopsis: Developmental processes like seed germination are characterised by massive transcriptome changes. This study compares seed transcriptome datasets of different Brassicaceae to identify stable expressed reference genes for cross-species qRT-PCR normalisation. A workflow is presented for improving RNA quality, qRT-PCR performance, and normalisation when analysing expression changes across species.
| Article in PDF format (1.2 MB) Supplemental data file (156 KB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |